Genomics/Epigenomics
Genomics/Epigenomics
165 - Potential Role of Khdc3 in Small RNA Inheritance and Dysregulation of Hepatic Gene Expression
Publication Number: 165.218

Liana Senaldi, MD (she/her/hers)
Neonatologist
Weill Cornell Medicine
New York, New York, United States
Presenting Author(s)
Background: The field of epigenetic inheritance is rapidly emerging and can potentially provide a mechanistic explanation for the many strongly inherited pediatric diseases that genomics-based approaches have largely been unable to explain. Organisms with mutations in the mammalian germ cell gene KHDC3L/Khdc3 have defective oocyte DNA methylation and abnormal fertility, yet the cellular and molecular function of this gene remains unknown. Previous research in our lab has found that oocytes from Khdc3-null mice have global dysregulation of small RNAs that regulate glucose and lipid metabolism. Given our data shows Khdc3 deficiency is associated with dysregulation of critical lipid and glucose metabolism genes, we next looked in the liver for evidence of metabolic dysfunction.
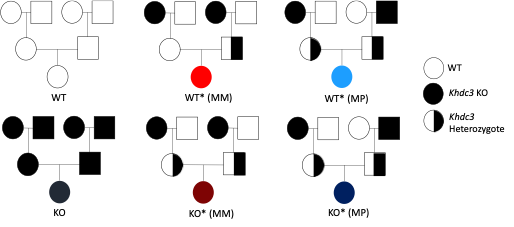
Objective: Evaluate global differences in hepatic gene expression in WT and Khdc3 knock-out (KO) mice, as well as WT mice generated from WT and Khdc3-null ancestry (WT* mice) and KO mice generated from WT and Khdc3-null ancestry (KO* mice).
Design/Methods:
Matings were designed to evaluate for differences in hepatic gene expression in genetically identical mice that have a different mutant ancestral history (Figure 1). RNA-sequencing was then performed on livers from WT, WT*, KO, and KO* mice to evaluate gene dysregulation.
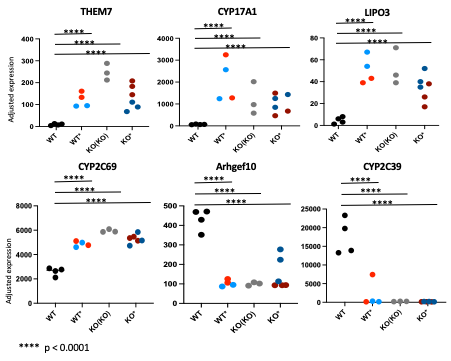
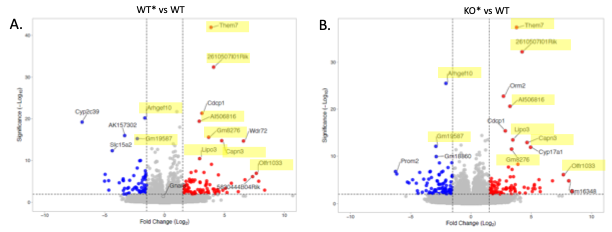
Results: Khdc3 KO mice had 1016 significantly dysregulated genes compared to WT mice (p< 0.05). Comparison of WT* mice with WT mice revealed dysregulated genes (p< 0.05) that significantly overlap with the dysregulated genes in the corresponding KO* mice generated from the same heterozygous parents, suggesting that abnormal gene expression may be driven by inherited epigenetic factors independent of direct genetic effects in the organism (Figure 2 & 3). Gene ontology enrichment for the genes uniquely dysregulated in WT* mice compared to WT mice showed genes involved in long chain fatty acid metabolism and lipid metabolism. Furthermore, WT** mice (generated from a WT* female mouse mated with a WT* male mouse) revealed that similar heptic gene dysregulation persisted across generations.
Conclusion(s):
Genetically identical mice with different Khdc3 mutation ancestry have different aberrantly expressed genes in their liver, suggesting that non-DNA molecules, such as small RNAs, are inherited at the time of fertilization that influence organ function. Ongoing experiments are aimed at examining small RNAs and DNA methylation in the liver.