Infectious Diseases
Infectious Diseases 1
626 - Single-cell RNA-sequencing reveals RSV- and SARS-CoV-2-specific cell states within the nasal mucosa of young children presenting to the emergency department
Saturday, April 29, 2023
3:30 PM - 6:00 PM ET
Poster Number: 626
Publication Number: 626.225
Publication Number: 626.225
Ying Tang, Boston Children's Hospital, Roxbury, MA, United States; Jaclyn M. Long, Boston Children's Hospital, Boston, MA, United States; Erica Langan, Boston Children's Hospital, Brookline, MA, United States; John L. Shultz, Boston Children's Hospital, Boston, MA, United States; Michelle Du, Boston Children's Hospital, Boston, MA, United States; Rebecca L. Wolf, Boston Children’s Hospital, Dennis Port, MA, United States; Joseph Griffiths, Boston Children's Hospital, Franklin, MA, United States; Tanya O. Robinson, University of Mississippi Medical Center, Jackson, MS, United States; Hannah R. Laird, University of Mississippi School of Medicine, Jackson, MS, United States; Neha S. Dhaliwal, University of Mississippi School of Medicine, Jackson, MS, United States; Anika Faruque, University of Mississippi School of Medicine, Madison, MS, United States; Sai Kota, University of Mississippi School of Medicine, Madison, MS, United States; Laura Wright-Sexton, University of Mississippi School of Medicine, Pascagoula, MS, United States; Thomas G. Wichman, University of Mississippi Medical Center, Jackson, MS, United States; Sarah C. Glover, University of Mississippi Medical Center, Jackson, MS, United States; Jose Ordovas-Montanes, Boston Children's Hospital, Boston, MA, United States; Bruce Horwitz, Boston Children's Hospital, Boston, MA, United States

Ying Tang, PhD (she/her/hers)
Research Fellow
Boston Children's Hospital
Roxbury, Massachusetts, United States
Presenting Author(s)
Background: Viral respiratory illnesses are a significant cause of morbidity in young children, who often lack prior immunity to common respiratory pathogens. RSV and now SARS-CoV-2 are common pathogens identified in young children presenting to the emergency department with acute respiratory symptoms, but whether these viruses induce distinct pathobiological processes in the respiratory mucosa of patients in this age group remains incompletely defined. We hypothesize that differences in cell tropism and the intrinsic anti-viral response of infected epithelial cells between these two viruses results in distinct host immune and inflammatory responses, the magnitude of which may associate with disease severity.
Objective: Our objective was to use single-cell RNA-sequencing (scRNA-seq) to identify the cellular targets of RSV and SARS-CoV-2 infection in the nasal mucosa of children less than two years of age, and compare the transcriptional response of infected epithelial cells, bystander epithelial cells, and immune cells to each virus at single cell resolution.
Design/Methods: We collected nasopharyngeal swabs from a convenience sample of 270 children less than two years of age presenting to a pediatric emergency department with signs and symptoms of acute viral respiratory infection, including 57 with RSV and 29 with SARS-CoV-2, as well as 32 age-matched controls presenting with minor traumatic injuries. We developed methodology to cryopreserve these swabs and subsequently perform scRNA-seq of isolated cells that allows simultaneous assessment of both host and viral transcriptomes at the single cell level.
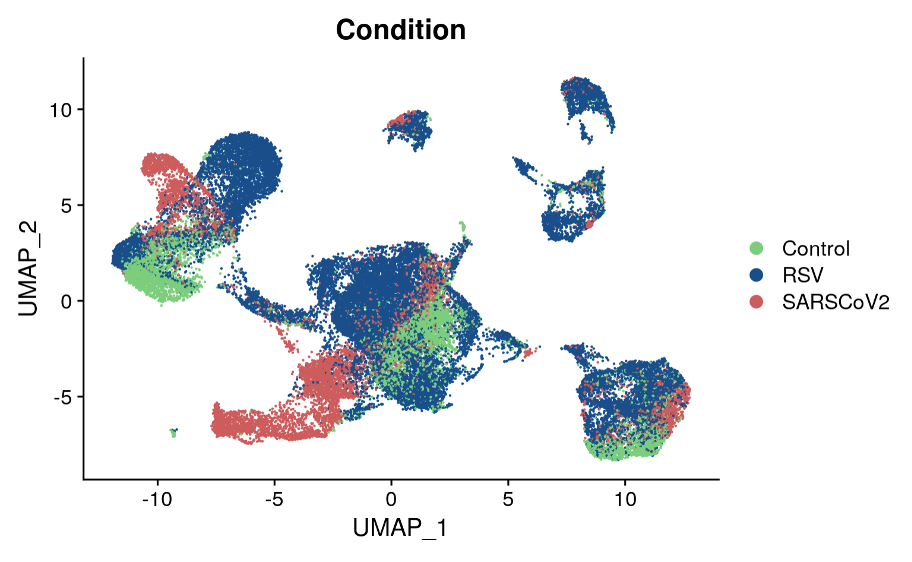
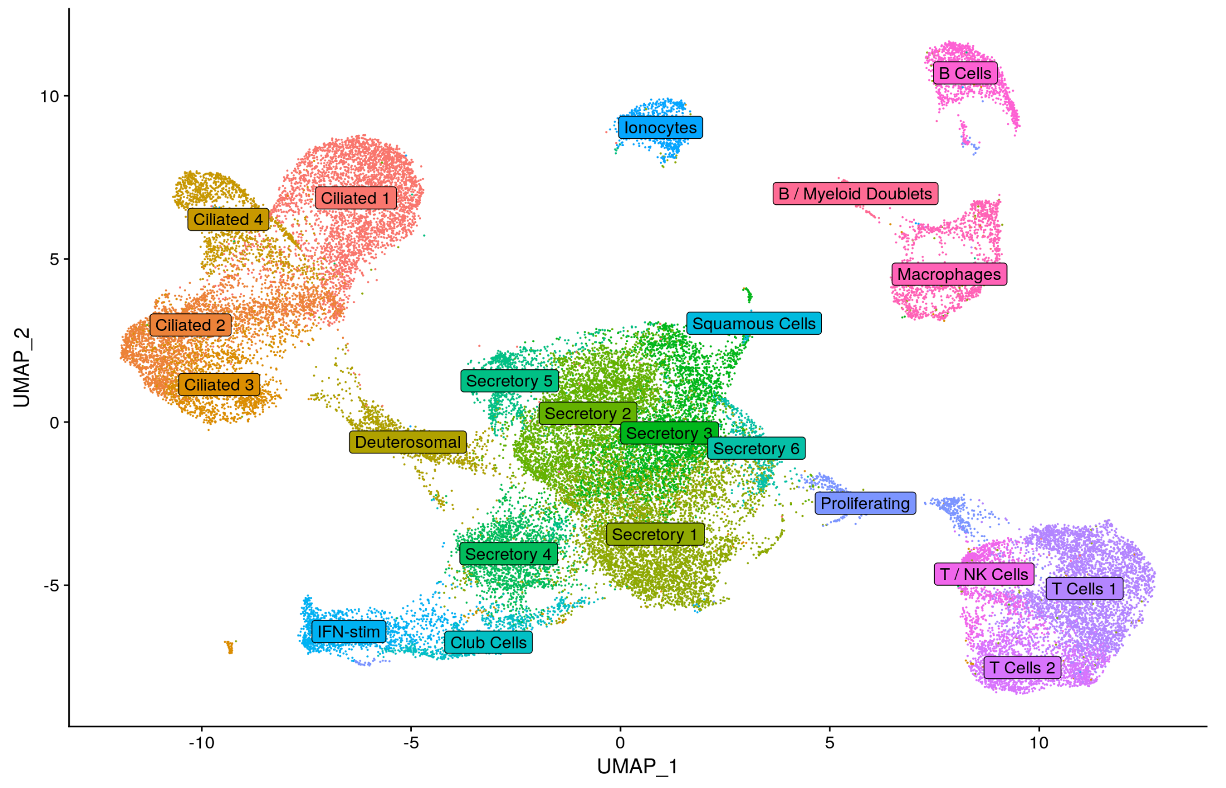
Results: Initially, we have sequenced and analyzed samples from 6 children infected with RSV, 6 infected with SARS-CoV-2, and 6 controls. We identified diverse populations of epithelial cells, immune cells, and ionocytes within the nasal mucosa of these young children. Consistent with our hypothesis, we have uncovered unique viral-specific cell states within subpopulations of ciliated and secretory epithelial cells that distinguish patients infected with each virus as well as controls, based in part on differential induction of anti-viral type I interferon responses.
Conclusion(s): Single-cell RNA sequencing of respiratory epithelial and immune cells collected from nasopharyngeal swabs can be used to characterize respiratory virus cell tropism, cell intrinsic and bystander epithelial cell responses to infection, and host immune responses with unprecedented cellular resolution. Ongoing analysis of this cohort could reveal viral-specific targets for the treatment of respiratory infections in young children.
.png)
