Telemedicine/EHR/Medical Informatics
Telemedicine/EHR/Medical Informatics 2
627 - Creation of clinically relevant clusters of neonates with machine learning
Monday, May 1, 2023
9:30 AM - 11:30 AM ET
Poster Number: 627
Publication Number: 627.455
Publication Number: 627.455
Emily Polidoro, The Mount Sinai Kravis Children's Hospital, New York, NY, United States; Girish Nadkarni, Icahn School of Medicine at Mount Sinai, New York, NY, United States; Jennifer Duchon, Mount Sinai, New York, NY, United States; Jessica Lewis, The Mount Sinai Kravis Children's Hospital, NEW YORK, NY, United States; Justin Kauffman, Icahn School of Medicine at Mount Sinai, Brooklyn, NY, United States

Emily Polidoro, MD (she/her/hers)
Fellow
The Mount Sinai Kravis Children's Hospital
New York, New York, United States
Presenting Author(s)
Background: Diseases in neonates often present with subtle and nonspecific findings. Automated processes that can aid clinicians in distinguishing disease states are desirable. Building machine learning models that make such predictions or classifications requires large labeled data sets, which is a time intensive manual process. A machine learning methodology that clusters neonates into clinically relevant groups provides weak bayesian priors for use in assigning classification probabilities, which is a first step in building a labeling model.
Objective: Generate clinically relevant clusters of neonates with unstructured EHR data as a first step in an automated labeling pipeline.
Design/Methods: EHR data including demographics, admission and discharge location, and medication administration was retrospectively obtained from two NICUs. An antibiotic course is defined as a continuous course of antibiotics, at least 48 hours long, with less than 48 hrs between doses. Topological analysis was performed on categorical type data such as admission/discharge type and antibiotic course composition in order to produce graph embeddings. Clinical variables (which also contain some categorical features) were embedded by factor analysis of mixed data for principal component analysis (FAMD-PCA). Semantic groupings were recovered from the clinical embeddings by maximizing normalized mutual information (NMI) over the first two principal components. NMI and markov clustering were used to find semantic groups over graph embeddings.
Results: There were 6961 courses of antibiotics identified in 5829 neonates. Recurrent patterns were identified among demographic data, admission/discharges, and antibiotic administrations leading to demonstration of 6, 9, and 9 clusters of patients respectively (Figure 1). The clusters were then examined for missingness and artifact; 1 of the clusters of demographic data was noted to be likely artifactual, so was excluded with minimal change in distributions. Each of the machine derived groups reflect clinically relevant phenotypic information. The 9 clusters of antibiotic combinations are characterized by a distinguishing antibiotic with intragroup variability preserved (Figure 2). The 5 remaining clusters of demographic data reflect commonly observed clinical patterns (Figure 3).
Conclusion(s): Machine learning can be utilized to distinguish patients into clinically relevant clusters, a first step to gathering a machine generated labeled data set.
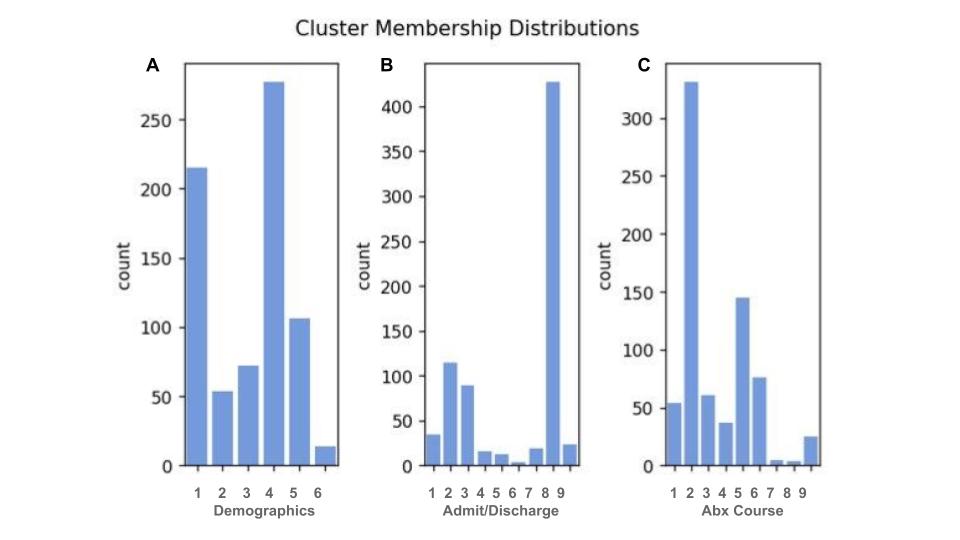
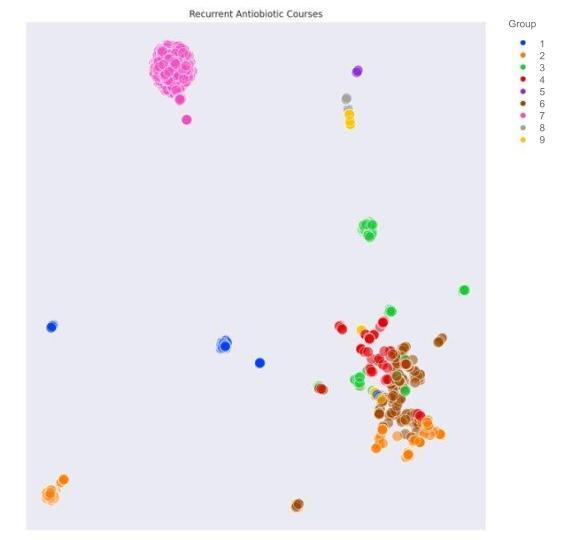
.jpg)
