General Pediatrics: All Areas
General Pediatrics 7
308 - Automated in-vivo placental segmentation using multimodal fusion based on deep neural networks
Publication Number: 308.408

Dhineshvikram Krishnamurthy, MS
Sr R&D Dev/Ops Engineer
Children's National Health System
Washington DC, District of Columbia, United States
Presenting Author(s)
Background:
Placental dysfunction is a leading cause of maternal pregnancy-related conditions such as hypertension, preeclampsia, and fetal growth restriction. However, there is often a delay in identifying placental disease due to a paucity of available tools that can accurately and noninvasively identify placental dysfunction early and in real time. Recent reports suggest quantitative MRI can improve accuracy and reproducibility in the structural, textural and morphometric analysis of placental development over traditional methods alone. However, precisely annotating the placental region in the presence of fetal and non-placental tissues is a complex, time-consuming, and labor-intensive process.
Objective:
To develop an automated segmentation method to improve reproducibility and accuracy in quantitative placental MR imaging.
Design/Methods:
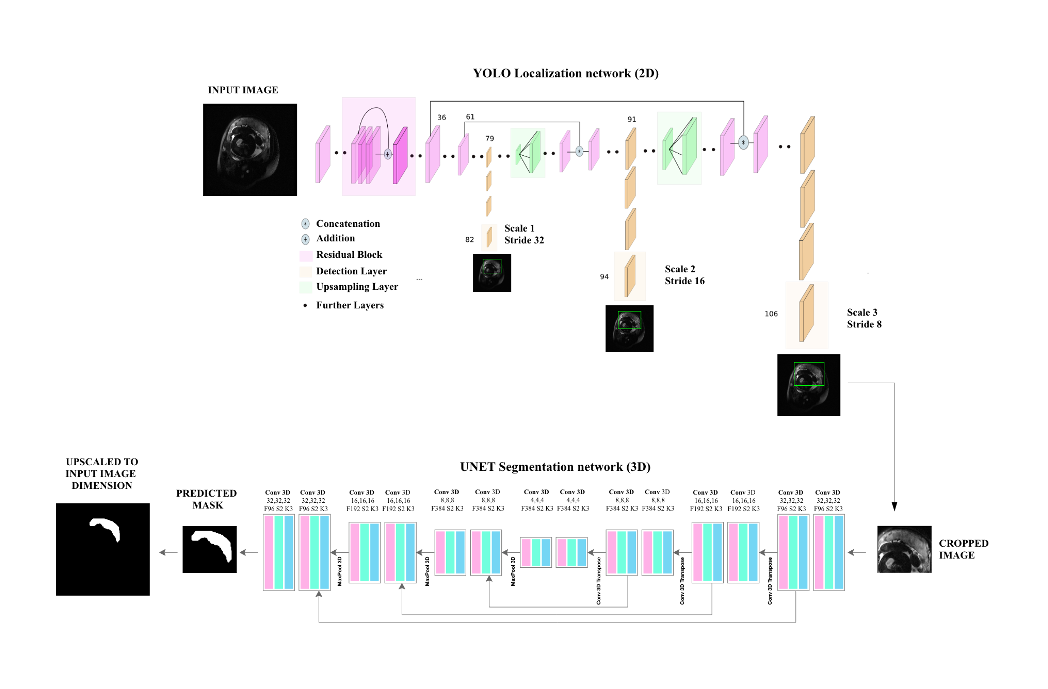
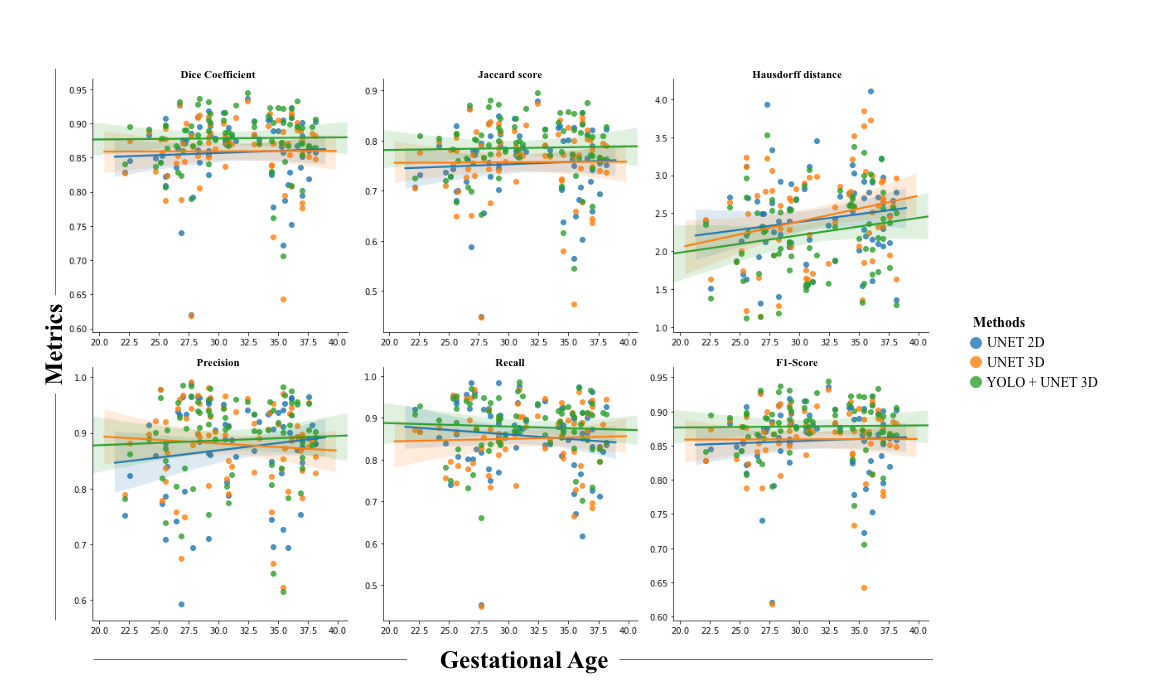
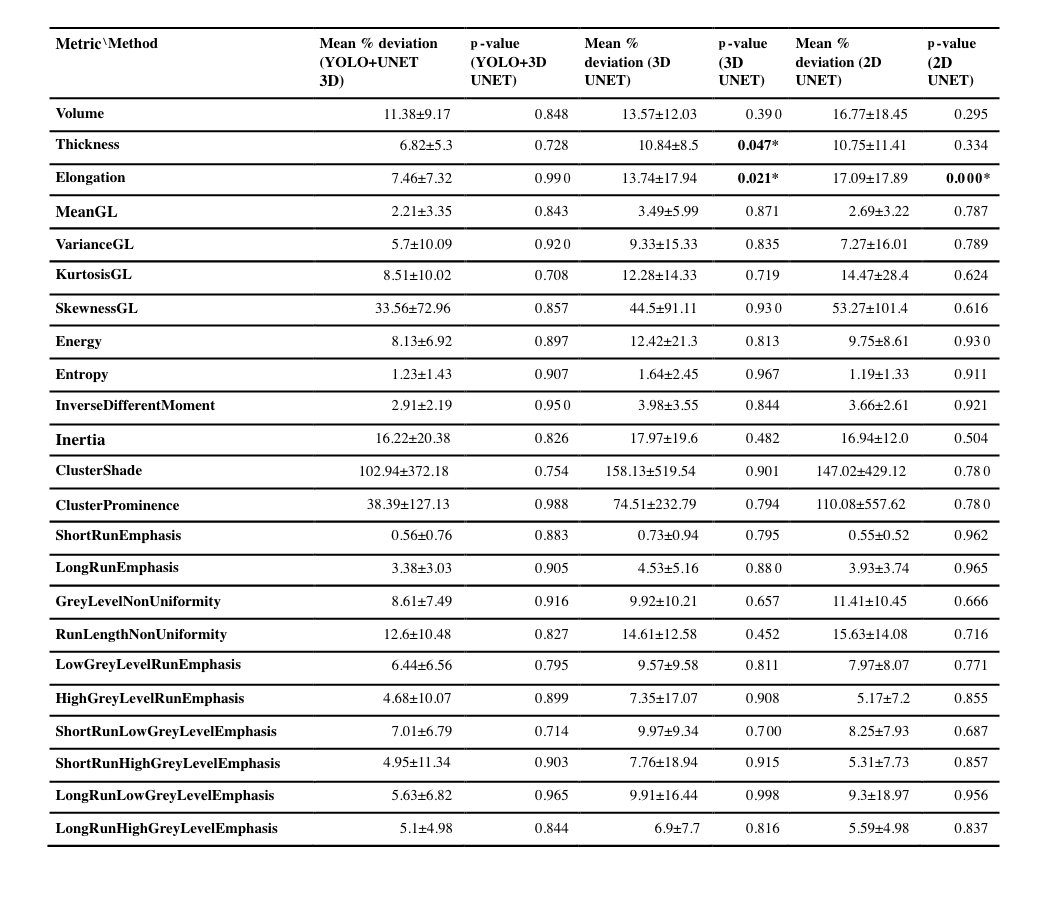
In this study, we propose an automatic segmentation method based on the fusion of two convolutional neural network models that perform close to the human annotator(Figure 1). The model was trained and evaluated on 360 fetal axial T2-weighted MRI studies obtained from 246 pregnant women ranging from 20-39 gestational weeks. Firstly, YOLO architecture based on a multi-scale loss function coarsely localized the placental region which reduced the complexity and mitigated class imbalance for the segmentation model. Secondly, Patch-based 3D-UNET an encode-decoder architecture finely segmented the placental region. Lastly, post-processing steps that included dilation, smoothing, slice filling, outlier rejection, and selecting the largest component were applied to achieve better performance. The proposed method achieved comparable performance to the human expert annotation at a mean dice score of 86.12%, a %volume difference of 9%, a precision of 91.62%, and a recall of 82.49%. The computation time of the proposed pipeline is ~40 seconds when run on a P100 NVIDIA GPU. This method outperformed 2D UNET and 3D UNET without YOLO using existing metrics reported in other research articles(Figure 2). The textural/morphometric measures of the proposed method showed improved performance relative to 2D UNET and 3D UNET without YOLO when compared to gold-standard manual segmentations(Table 1).
Results:
Conclusion(s):
The proposed method is fast, reliable, robust, and efficient in segmenting placental regions precisely, which will be helpful in evaluating placental structural development to identify early biomarkers of risk to maternal-fetal wellness. Since this method runs in near real-time, it has the potential to be deployed in a clinical setting for human placental image analysis.