Neonatal GI Physiology & NEC
Neonatal GI Physiology & NEC 2: Gut and Liver Health
344 - Exclusionary Klebsiella Species Competition in Preterm Infants at Risk for Necrotizing Enterocolitis
Publication Number: 344.134

Adam P. Matson, MD, MSc (he/him/his)
Attending Neonatologist, Physician Scientist
University of Connecticut School of Medicine
Farmington, Connecticut, United States
Presenting Author(s)
Background: Necrotizing enterocolitis (NEC) is a devastating intestinal disorder of premature infants that involves abnormal gut microbiota triggering an overly exuberant pro-inflammatory response. Whether specific bacteria contribute to dysbiotic microbial profiles in NEC remains an open question. Klebsiella has been implicated in previous studies, but methods of analysis usually failed to discriminate Klebsiella species.
Objective:
To differentiate Klebsiella species and strains in the gut microbiome of preterm infants with and without NEC.
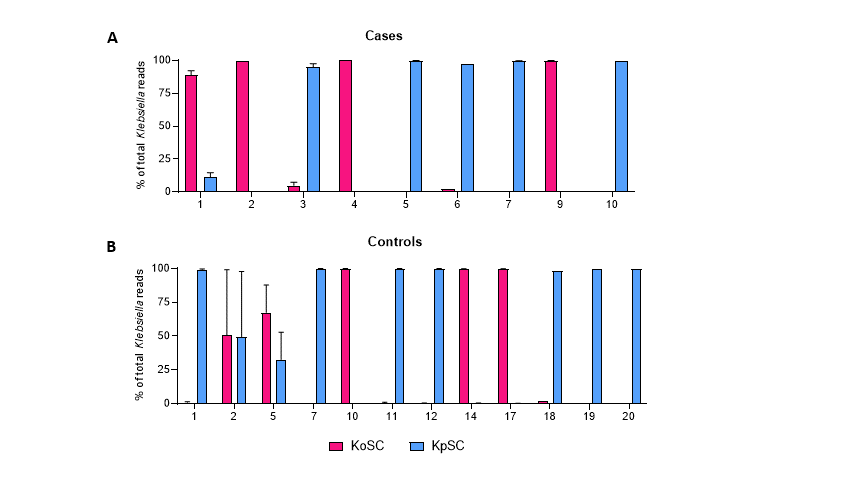
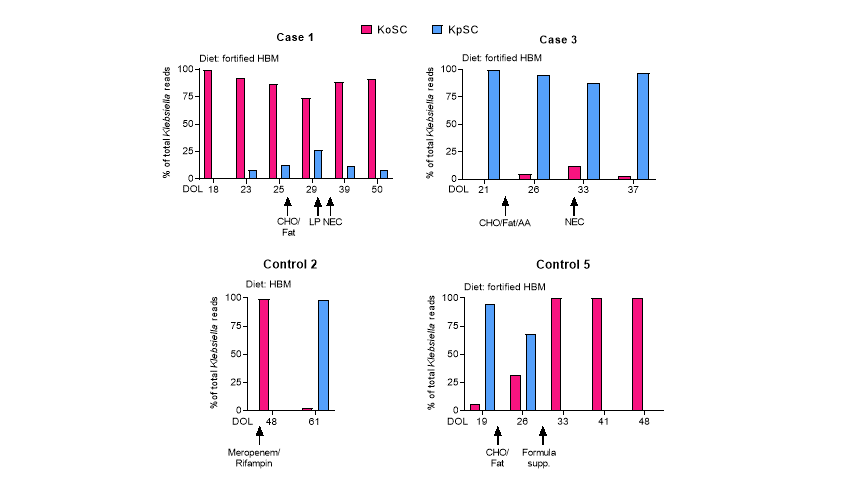
Design/Methods: A novel ~2,500 base rRNA amplicon spanning the bacterial 16S and 23S rRNA genes was used to generate amplicon sequence variant (ASV) fingerprints for Klebsiella oxytoca and Klebsiella pneumoniae species complexes (KoSC and KpSC, respectively) and co-occurring fecal bacterial strains from 10 preterm infants with NEC and 20 matched controls. Complementary approaches were used to identify cytotoxin-producing isolates of KoSC.
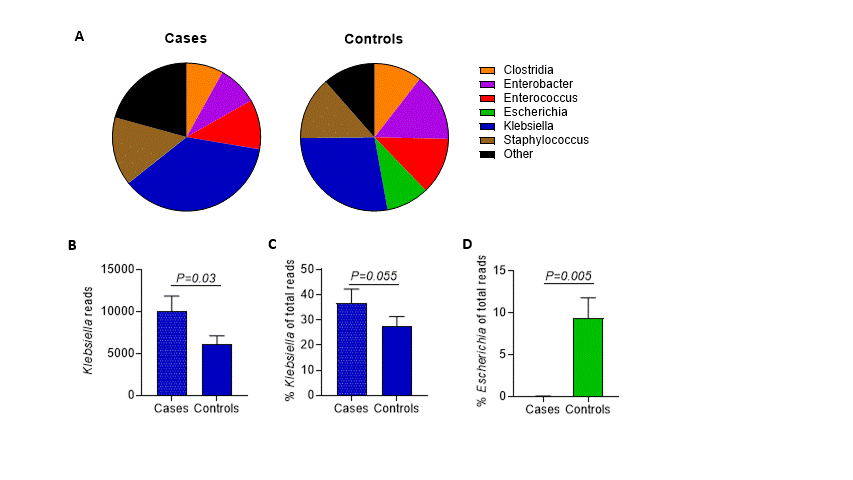
Results: Klebsiella species colonized most preterm infants, were more prevalent in NEC subjects versus controls, and replaced Escherichia in NEC subjects. Single KoSC or KpSC ASV fingerprinted strains dominated the gut microbiota, suggesting exclusionary Klebsiella competition for luminal resources. Enterococcus faecalis was co-dominant with KoSC but present infrequently with KpSC. Cytotoxin-producing KoSC members were identified in most NEC subjects and were less frequent in controls. Few Klebsiella strains were shared between subjects.
Conclusion(s): ASV fingerprinting enabled differentiation of KoSC and KpSC strains in gut microbiomes of premature infants. Inter-species Klebsiella competition, within an environment of KoSC and E. faecalis cooperation, appears to be an important factor for the development of NEC. Preterm infants seem to acquire Klebsiella primarily through routes other than patient-to-patient transmission. Monitoring and control of competitive mechanisms and elucidation of NEC strain acquisition could be used to limit NEC.