Emergency Medicine: All Areas
Emergency Medicine 1
17 - High-Throughput Sequencing to Identify Bacteremia in Children with Cancer
Friday, April 28, 2023
5:15 PM - 7:15 PM ET
Poster Number: 17
Publication Number: 17.11
Publication Number: 17.11
Son H. McLaren, Columbia University Vagelos College of Physicians and Surgeons, New York, NY, United States; Nischay Mishra, Columbia University, New york, NY, United States; Nobuko Hijiya, Columbia University Vagelos College of Physicians and Surgeons, new york, NY, United States; James Ng, Columbia University, BRONX, NY, United States; Cheng Guo, columbia university, new york, NY, United States; Susan Whittier, Columbia University Vagelos College of Physicians and Surgeons, Cape Coral, FL, United States; Erica Mariani, NYU Bellevue, NY, NY, United States; Laura Glaser, New York Presbyterian Hospital, Fort Lee, NJ, United States; Irene Frantzis, Columbia University Vagelos College of Physicians and Surgeons, New York, NY, United States; Marc T. Vindas, Rutgers Cancer Institute of New Jersey, West New York, NJ, United States; Cheng-Shiun Leu, Columbia University, New York, NY, United States; Peter Dayan, Columbia University, Hastings on Hudson, NY, United States
.jpg)
Son H. McLaren, MD, MS (she/her/hers)
Assistant Professor of Pediatrics (in Emergency Medicine)
Columbia University Vagelos College of Physicians and Surgeons
New York, New York, United States
Presenting Author(s)
Background: Bacteremia is a major source of morbidity and mortality in children with cancer; however, current methods to accurately and efficiently diagnose bacteremia are limited.
Objective: We aimed to determine the concordance between a novel high-throughput sequencing (HTS) platform to detect bacteremia called BacCapSeq and the reference standard blood culture.
Design/Methods: In this single center prospective cross-sectional pilot study, we enrolled children ≤19 years old with cancer presenting to the oncology clinic or the emergency department with fever (temperature ≥38.0ºC) and for whom a blood culture was obtained. Afebrile controls (children 5-19 years old with cancer but no fever, and asymptomatic otherwise healthy adult volunteers >19 years old) were also enrolled to assess detection of bacteria by HTS in otherwise clinically well children and adults in whom bacteremia would not be expected. In addition to standardized history, exam, and 2-week telephone follow up, blood samples were collected for each patient for HTS. Analysis of sequencing data generated by BacCapSeq was performed using two bioinformatics pipelines (Kraken2 and MetaPhlAn2). For febrile children, we calculated concordance between HTS and blood culture results using percent agreement. We defined positive agreement as HTS identifying all bacteria in a blood culture, irrespective of whether HTS identified additional bacteria judged unlikely to be clinically relevant. Clinical relevance of discordant organisms was determined by two infectious disease physicians through medical record review.
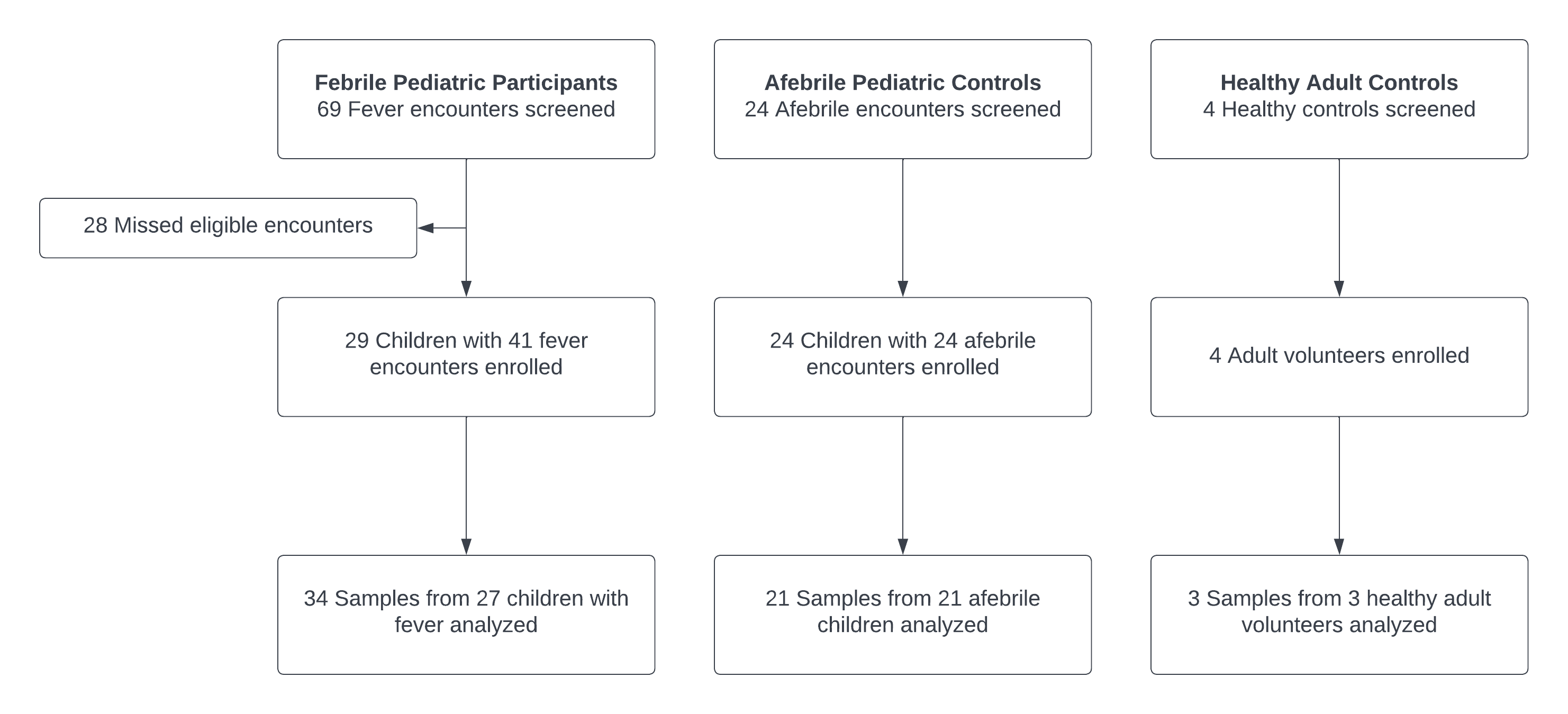
Results: We enrolled 29 children with 41 febrile encounters, 24 children with 24 afebrile encounters, and 4 healthy adult controls. Of these, samples from 34 febrile encounters, 21 afebrile encounters, and 3 adult controls were analyzed (Figure). Twenty-two of the febrile encounters (64.7%) resulted in hospital admissions and 2/22 (9.0%) in death. Five febrile patients had bacteremia based on blood culture results. Positive percent agreement (PPA) and negative percent agreement (NPA) between blood culture and HTS data analyzed by MetaPhlAn2 was 60% (95% CI 14.7-94.7%) and 100% (95% CI 88.1-100.0%), respectively (Table 1). When analyzed using Kraken2, PPA and NPA were 40% (95% ci 5.3-85.3%) and 97% (95% CI 82.2-99.9%), respectively. Discordant organisms included those with unlikely or potential clinical relevance (Table 2). HTS identified bacteria without clinical relevance in afebrile controls.
Conclusion(s): HTS had modest concordance with blood culture. Further refinement and clinical validation are needed prior to its use in the clinical setting.
.png)
.jpg)
